1. 所有訂購之細菌株均為96 well
plate format。
2. 細菌株均為15% glycerol
stock,體積約為100μl / well。
3. 處理程序:回溫→離心(2000rpm, 1min)→撕封膜。
4. 請以streak out
的方式挑選單一菌落,經16小時培養之後,單一菌落直徑約為0.5mm大小, 請勿挑選培養盤上菌落較大者作液體培養,以防選到變異株;
每一clone至少挑選2顆菌落,之後萃取質體,並做定序 (註一)。
若塗佈養菌失敗,可先從stock進行少量液體培養後,再以streak out方式挑菌。
5.
本細菌庫所使用之質體為pLKO.1(Amp+),生長速度較慢,故建議液體培養時請使用Terrific Broth (TB,註二)。
*註一:
pLKO.1-puro轉移載體以U6 promoter表達shRNA,本核心設施使用以下primers其中之一定序之:
(1) LKO_shRNA/F (forward primer):
5’-acaaaatacgtgacgtag-3’ (for sequencing forward strand of shRNA)。
(2) LKO_shRNA/R (reverse primer):
5’-ctgttgctattatgtctac-3’ (for sequencing reverse strand of shRNA)。
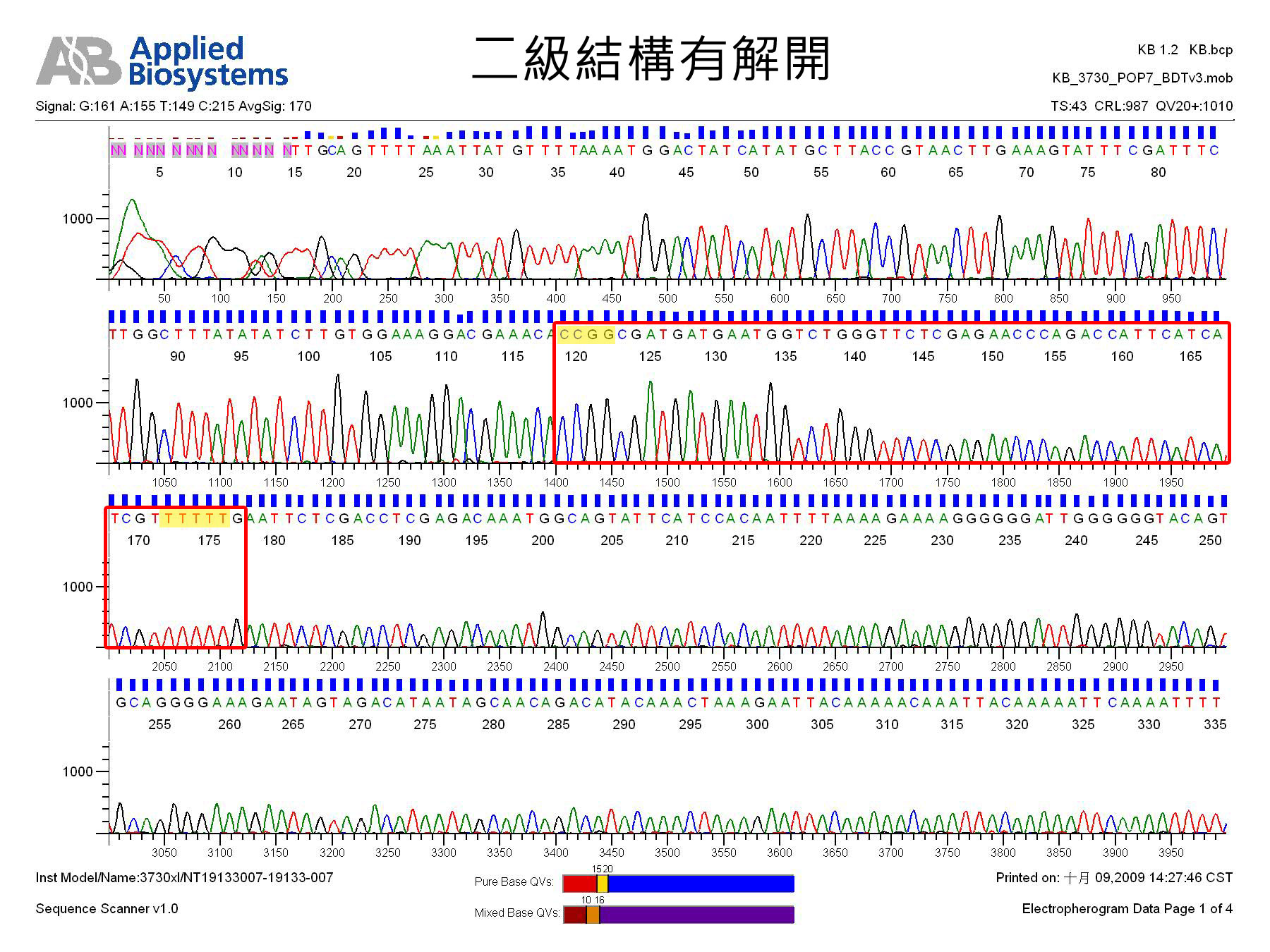
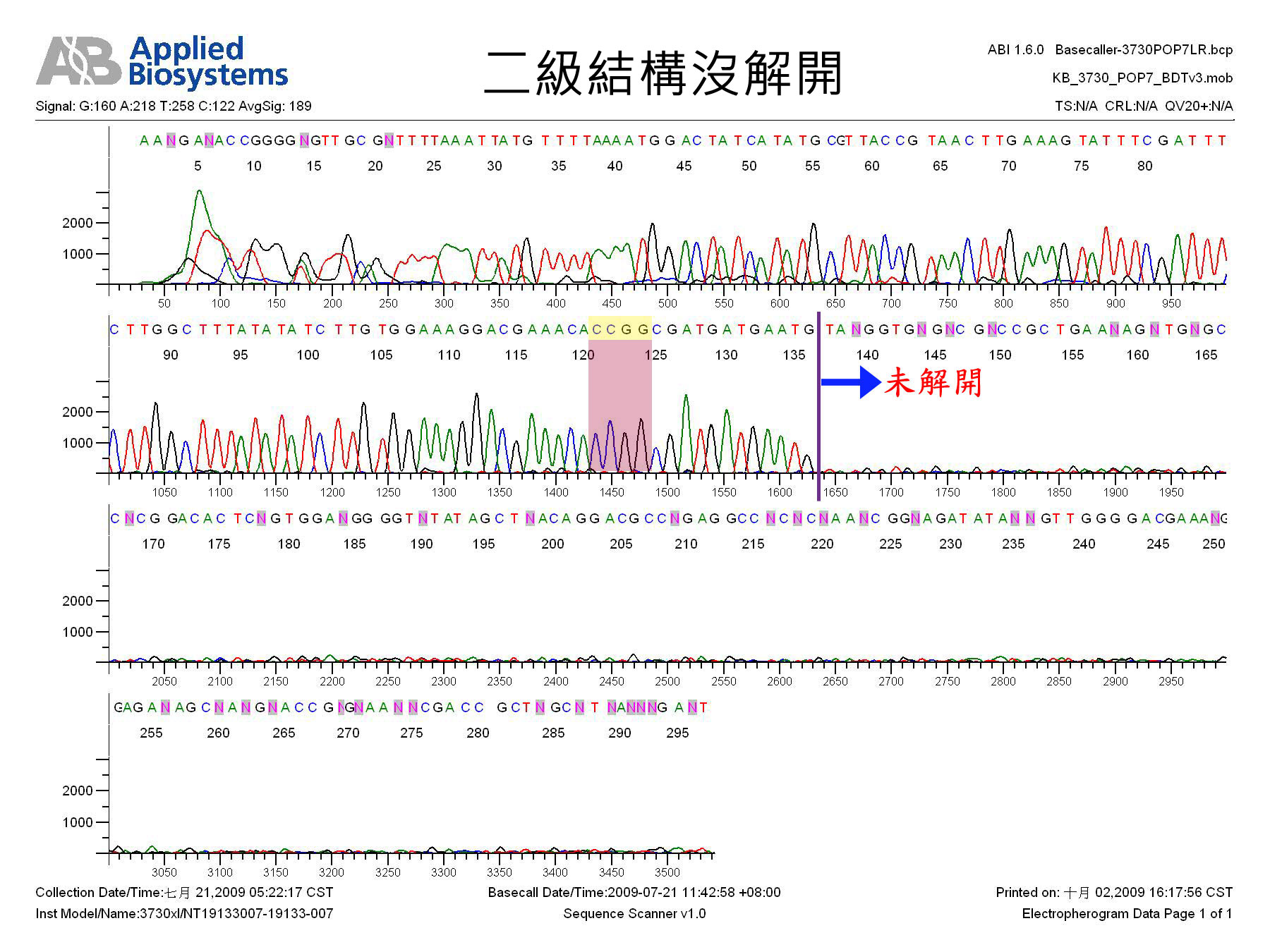
shRNA因有二級結構,本核心設施建議定序方法如下:
(1) 請在樣品中加入5% ~10% DMSO,定序之;或
(2) 請定序中心以dGTP BigDye V1.1 kit或類似產品定序之。
 |
 |
| 二級結構有解開(圖片按下去可放大) |
二級結構沒解開(圖片按下去可放大) |
*註二: Terrific Broth 配方,正確配法請參考 Molecular cloning。
| Tryptone |
12g/L |
| Yeast extract |
24g/L |
| Glycerol(100%) |
4ml/L |
| KH2PO4 |
2.31g/L |
| K2HPO4 |
12.54g/L |
Ampicillin or
Carbenicillin final concentration:100mg / L
# 當您的實驗室開始使用細菌株進行實驗後,如果有【定序】、【Knockdown結果】或【論文發表】時,請feedback至本核心設施,俾使整合shRNA
database或將成果(論文)呈報到MOST,謝謝合作 #
1. C6-1-1 product is contained in 96-well plate format with 100μL volume for each clone.
2. Bacteria (DH5a) are frozen in 15% glycerol stock.
3. Recover the plate in room temperature → centrifuge (2000rpm, 1min) → unseal the plate → mix the bacteria glycerol stock gently → take 3-5μL to Luria broth (LB/Amp+) → incubate bacterial culture at 37°C for 12-18 hr in a shaking incubator.
4. Obtain the bacteria culture with loop/ tip → gently spreak the bacteria over the LB agar plate(Amp+) → incubate LB agar plate at 37°C for 12-18 hr → pick 2-3 single colonies to Luria broth (LB/+amp) and culture at 37°C o/n → plasmid DNA extraction → sequencing check.
5. If plate or broth inoculated from C6 Core bacterial stock shows no growth after incubation or unexpected sequencing data, please give us feedback and we’ll replace the new clone for free.
*Sequencing primer (use either one only):
(1) LKO_shRNA/F (forward primer):
5’-acaaaatacgtgacgtag-3’ (for sequencing forward strand of shRNA)。
(2) LKO_shRNA/R (reverse primer):
5’-ctgttgctattatgtctac-3’ (for sequencing reverse strand of shRNA)。
Protocol for sequencing through the secondary structure of the shRNA hairpin structure:
(1) Add 5%-10% DMSO to each sample.
(2) Remind sequencing center of the secondary structure sample.
 |
 |
| Secondary structure is resolved. |
Secondary structure problem. |
*A single colony should look like a white dot growing on the solid medium
*pLKO.1(Amp+) is low copy plasmid number plasmid. You can use Terrific Broth (TB) to improve the yield of plasmid DNA from E. coli.
| Tryptone |
12g/L |
| Yeast extract |
24g/L |
| Glycerol(100%) |
4ml/L |
| KH2PO4 |
2.31g/L |
| K2HPO4 |
12.54g/L |
Ampicillin or
Carbenicillin final concentration:100mg / L
*Please send us feedback when the knock down efficiency data is completed or the results are published in journal, C6 core will collect the results to MOST, thank you.